|
|
Research Themes:
We believe in a border less world of scientific knowledge. Our research aims to excel in the technical, biological and applied aspects of bioinformatics. Despite expanding horizontally in several areas, we hate being superficial and repetitive without innovation. Thus, we aim to build a team of researchers with genuine passion in science, trying to unravel solutions with insight, rigour and utmost honesty and sincerity.
1. Core Bioinformatics (Basic Research)
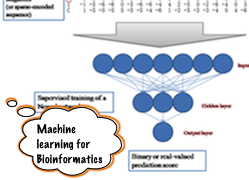
Development of sequence-based methods to understand and predict Biomolecular interactions in general and protein-DNA interactions in particular. We approach the problem from an integrated data-driven perspective taking cues from physics and informatics. Machine learning methods in general and neural networks in particular are our choice of technique and we often hop on to developing totally new training algorithm or neural network architecture while solving our problems.
2. Applied and Collaborative Research
Our recent interests in applied research are related to topics in systems biology. This includes providing predictive and molecular basis of gene expression and its regulation by transcription factors, miRNA and proteins. We have been working on novel methods of cross-experiment data processing, coherent themes in gene sets for enrichment analysis and tissue-specificity of biological pathways. Our current collaborative projects involve strain-specificity analysis of microbial genomes, condition-specific functional annotations and de novo genome assembly.
3. Tools and web servers
We have developed more than a dozen bioinformatics tools related to my research topics. Most popular among them are sequence-based method to predict binding sites, solvent accessibility and most recently DNA conformational dynamics.
Research Topics
 |
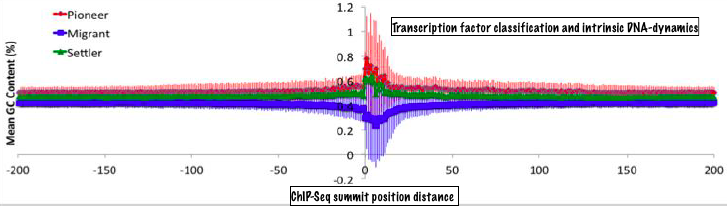
Sequence to interactions by machine learningOur bioinformatics research has focussed on understanding protein-DNA (and to a smaller extent other biomolecular) interactions with an ultimate objective of predicting them from sequence. To this end, we have studied large scale data sets from protein-DNA complexes, ChIP-Seq and DNAse-seq experiments, developed machine learning tools to predict DNA-binding residues and proteins, dissected thermodynamics and evolutionary patterns of protein-DNA interactions. These results have been announced as a number of research papers, which have received due attention by scientific community evident from citation record. |
|
|
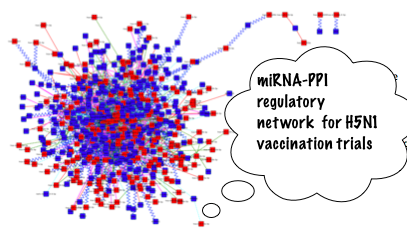
Systems Biology and transcriptome analysisOur more recent interests are related to systems biology track of bioinformatics research. Primary aim of this research is to provide a molecular basis of gene expression and thereby provide predictive and testable hypothesis about biological outcomes such as disease, cell-fate and adverse effects of vaccination. |
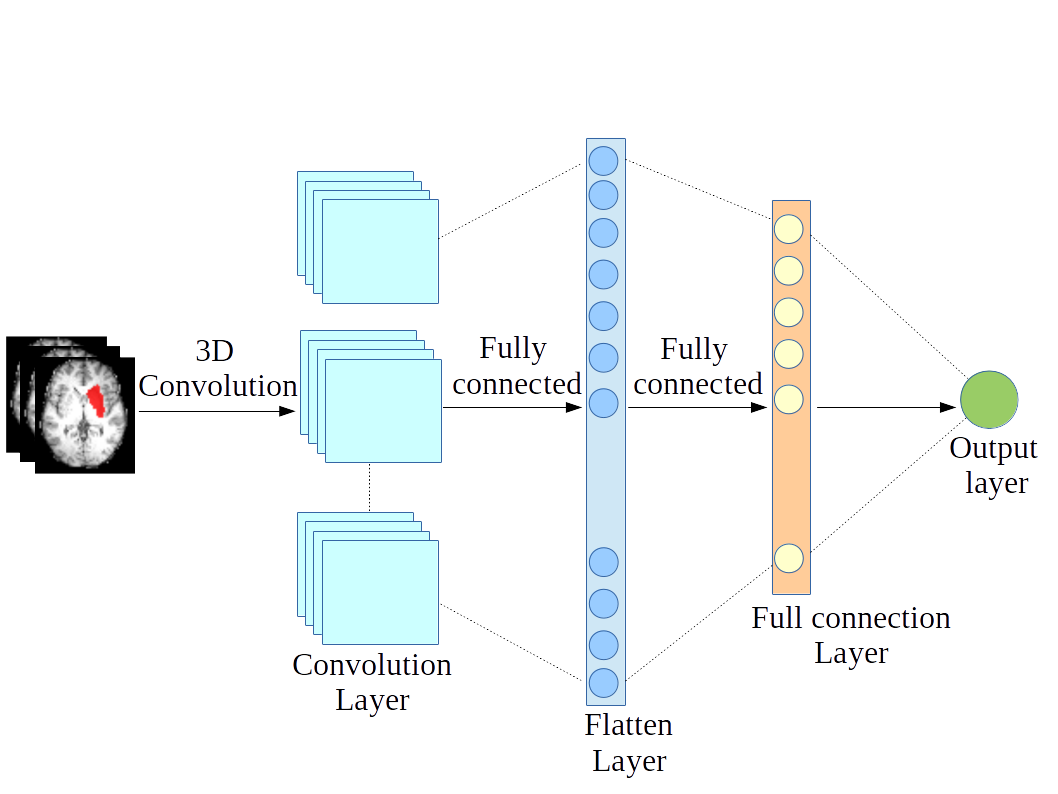 |
Deep learning for predicting cognitive performanceOptimal use of contrast imaging techniques to assess, diagnose and interpret behavioral and motor disorders caused by brain injuries is a subject of intensive research. The problem of language deficit prediction from MRI data has been recently addressed. The goal of the study is to gain insights into the structural patterns of lesions, which lead to language deficiency. There is no way to find the best predictive computational model, so it needs a separate more focussed treatment. |